
During my Ph.D., I created tools for people who write programs to obtain insights from data. Millions of professionals in fields ranging from science, engineering, business, finance, public policy, and journalism, as well as numerous students and computer hobbyists, all perform this sort of programming on a daily basis.
Shortly after I wrote my dissertation in 2012, the term Data Science started appearing everywhere. Some industry pundits call data science the “sexiest job of the 21st century.” And universities are putting tremendous funding into new Data Science Institutes.
I now realize that data scientists are one main target audience for the tools that I created throughout my Ph.D. However, that job title was not as prominent back when I was in grad school, so I didn’t mention it explicitly in my dissertation.
What do data scientists do at work, and what challenges do they face?
This post provides an overview of the modern data science workflow, adapted from Chapter 2 of my Ph.D. dissertation, Software Tools to Facilitate Research Programming.
The Data Science Workflow
The figure below shows the steps involved in a typical data science workflow. There are four main phases, shown in the dotted-line boxes: preparation of the data, alternating between running the analysis and reflection to interpret the outputs, and finally dissemination of results in the form of written reports and/or executable code.
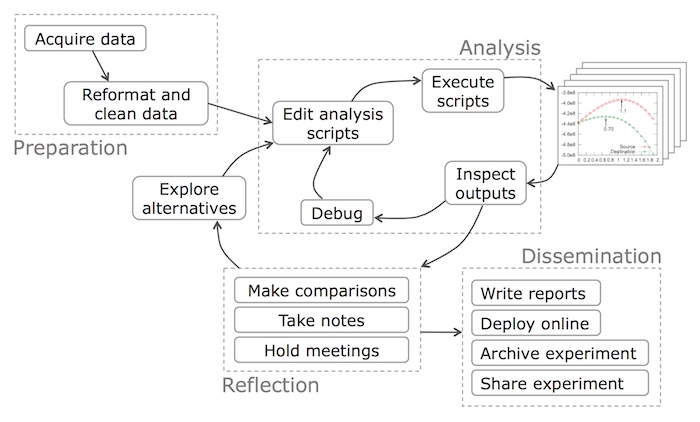
Preparation Phase
Before any analysis can be done, the programmer (data scientist) must first acquire the data and then reformat it into a form that is amenable to computation.
Acquire data: The obvious first step in any data science workflow is to acquire the data to analyze. Data can be acquired from a variety of sources. e.g.,:
- Data files can be downloaded from online repositories such as public websites (e.g., U.S. Census data sets).
- Data can be streamed on-demand from online sources via an API (e.g., the Bloomberg financial data stream).
- Data can be automatically generated by physical apparatus, such as scientific lab equipment attached to computers.
- Data can be generated by computer software, such as logs from a webserver or classifications produced by a machine learning algorithm.
- Data can be manually entered into a spreadsheet or text file by a human.
The main problem that programmers face in data acquisition is keeping track of provenance, i.e., where each piece of data comes from and whether it is still up-to-date. It is important to accurately track provenance, since data often needs to be re-acquired in the future to run updated experiments. Re-acquisition can occur either when the original data sources get updated or when researchers want to test alternate hypotheses. Also, provenance can enable downstream analysis errors to be traced back to the original data sources.
Data management is a related problem: Programmers must assign names to data files that they create or download and then organize those files into directories. When they create or download new versions of those files, they must make sure to assign proper filenames to all versions and keep track of their differences. For instance, scientific lab equipment can generate hundreds or thousands of data files that scientists must name and organize before running computational analyses on them.
A secondary problem in data acquisition is storage: Sometimes there is so much data that it cannot fit on a single hard drive, so it must be stored on remote servers. However, anecdotes and empirical studies indicate that a significant amount of data analysis is still done on desktop machines with data sets that fit on modern hard drives (i.e., less than a terabyte).
Reformat and clean data: Raw data is probably not in a convenient format for a programmer to run a particular analysis, often due to the simple reason that it was formatted by somebody else without that programmer’s analysis in mind. A related problem is that raw data often contains semantic errors, missing entries, or inconsistent formatting, so it needs to be “cleaned” prior to analysis.
Programmers reformat and clean data either by writing scripts or by manually editing data in, say, a spreadsheet. Many of the scientists I interviewed for my dissertation work complained that these tasks are the most tedious and time-consuming parts of their workflow, since they are unavoidable chores that yield no new insights. However, the chore of data reformatting and cleaning can lend insights into what assumptions are safe to make about the data, what idiosyncrasies exist in the collection process, and what models and analyses are appropriate to apply.
Data integration is a related challenge in this phase. For example, Christian Bird, an empirical software engineering researcher that I interviewed at Microsoft Research, obtains raw data from a variety of .csv and XML files, queries to software version control systems and bug databases, and features parsed from an email corpus. He integrates all of these data sources together into a central MySQL relational database, which serves as the master data source for his analyses.
In closing, the following excerpt from the introduction of the book Python Scripting for Computational Science summarizes the extent of data preparation chores:
Scientific Computing Is More Than Number Crunching: Many computational scientists work with their own numerical software development and realize that much of the work is not only writing computationally intensive number-crunching loops. Very often programming is about shuffling data in and out of different tools, converting one data format to another, extracting numerical data from a text, and administering numerical experiments involving a large number of data files and directories. Such tasks are much faster to accomplish in a language like Python than in Fortran, C, C++, C#, or Java.
In sum, data munging and organization are human productivity bottlenecks that must be overcome before actual substantive analysis can be done.
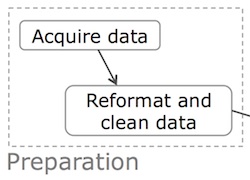
Analysis Phase
The core activity of data science is the analysis phase: writing, executing, and refining computer programs to analyze and obtain insights from data. I will refer to these kinds of programs as data analysis scripts, since data scientists often prefer to use interpreted “scripting” languages such as Python, Perl, R, and MATLAB. However, they also use compiled languages such as C, C++, and Fortran when appropriate.
The figure below shows that in the analysis phase, the programmer engages in a repeated iteration cycle of editing scripts, executing to produce output files, inspecting the output files to gain insights and discover mistakes, debugging, and re-editing.
The faster the programmer can make it through each iteration, the more insights can potentially be obtained per unit time. There are three main sources of slowdowns:
- Absolute running times: Scripts might take a long time to terminate, either due to large amounts of data being processed or the algorithms being slow, which could itself be due to asymptotic “Big-O” slowness and/or the implementations being slow.
- Incremental running times: Scripts might take a long time to terminate after minor incremental code edits done while iterating on analyses, which wastes time re-computing almost the same results as previous runs.
- Crashes from errors: Scripts might crash prematurely due to errors in either the code or inconsistencies in data sets. Programmers often need to endure several rounds of debugging and fixing banal bugs such as data parsing errors before their scripts can terminate with useful results.
File and metadata management is another challenge in the analysis phase. Repeatedly editing and executing scripts while iterating on experiments causes the production of numerous output files, such as intermediate data, textual reports, tables, and graphical visualizations. For example, the figure below
shows a directory listing from a computational biologist’s machine that contains hundreds of PNG output image files, each with a long and cryptic filename. To track provenance, data scientists often encode metadata such as version numbers, script parameter values, and even short notes into their output filenames. This habit is prevalent since it is the easiest way to ensure that metadata stays attached to the file and remains highly visible. However, doing so leads to data management problems due to the abundance of files and the fact that programmers often later forget their own ad-hoc naming conventions. The following email snippet from a Ph.D. student in bioinformatics summarizes these sorts of data management woes:
Often, you really don’t know what’ll work, so you try a program with a combination of parameters, and a combination of input files. And so you end up with a massive proliferation of output files. You have to remember to name the files differently, or write out the parameters every time. Plus, you’re constantly tweaking the program, so the older runs may not even record the parameters that you put in later for greater control. Going back to something I did just three months ago, I often find out I have absolutely no idea what the output files mean, and end up having to repeat it to figure it out.
Lastly, data scientists do not write code in a vacuum: As they iterate on their scripts, they often consult resources such as documentation websites, API usage examples, sample code snippets from online forums, PDF documents of related research papers, and relevant code obtained from colleagues.
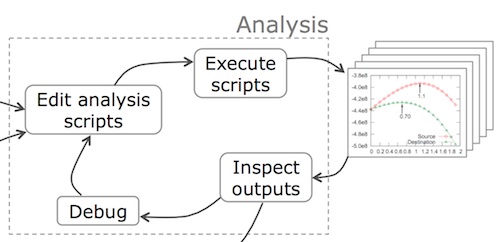
Reflection Phase
Data scientists frequently alternate between the analysis and reflection phases while they work, as denoted by the arrows between the two respective phases in the figure below:
Whereas the analysis phase involves programming, the reflection phase involves thinking and communicating about the outputs of analyses. After inspecting a set of output files, a data scientist might perform the following types of reflection:
Take notes: People take notes throughout their experiments in both physical and digital formats. Physical notes are usually written in a lab notebook, on sheets of looseleaf paper, or on a whiteboard. Digital notes are usually written in plain text files, “sticky notes” desktop widgets, Microsoft PowerPoint documents for multimedia content, or specialized electronic notetaking applications such as Evernote or Microsoft OneNote. Each format has its advantages: It is often easier to draw freehand sketches and equations on paper, while it is easier to copy-and-paste programming commands and digital images into electronic notes. Since notes are a form of data, the usual data management problems arise in notetaking, most notably how to organize notes and link them with the context in which they were originally written.
Hold meetings: People meet with colleagues to discuss results and to plan next steps in their analyses. For example, a computational science Ph.D. student might meet with her research advisor every week to show the latest graphs generated by her analysis scripts. The inputs to meetings include printouts of data visualizations and status reports, which form the basis for discussion. The outputs of meetings are new to-do list items for meeting attendees. For example, during a summer internship at Microsoft Research working on a data-driven study of what factors cause software bugs to be fixed, I had daily meetings with my supervisor, Tom Zimmermann. Upon inspecting the charts and tables that my analyses generated each day, he often asked me to adjust my scripts or to fork my analyses to explore multiple alternative hypotheses (e.g., “Please explore the effects of employee location on bug fix rates by re-running your analysis separately for each country.”).
Make comparisons and explore alternatives: The reflection activities that tie most closely with the analysis phase are making comparisons between output variants and then exploring alternatives by adjusting script code and/or execution parameters. Data scientists often open several output graph files side-by-side on their monitors to visually compare and contrast their characteristics. Diana MacLean observed the following behavior in her shadowing of scientists at Harvard:
Much of the analysis process is trial-and-error: a scientist will run tests, graph the output, rerun them, graph the output, etc. The scientists rely heavily on graphs — they graph the output and distributions of their tests, they graph the sequenced genomes next to other, existing sequences.
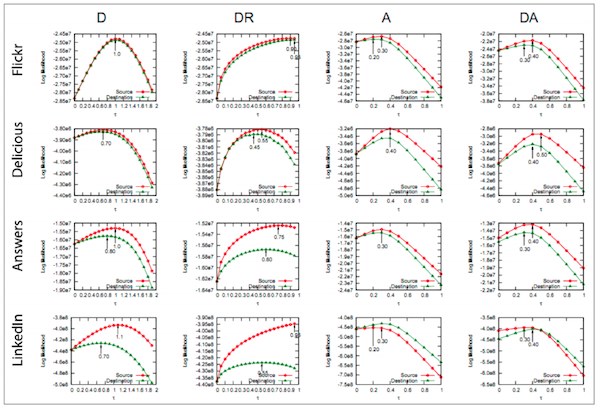
The figure below shows an example set of graphs from social network analysis research, where four variants of a model algorithm are tested on four different input data sets:
This example is the final result from a published paper and Ph.D. dissertation; during the course of running analyses, many more of these types of graphs are produced by analysis scripts. Data scientists must organize, manage, and compare these graphs to gain insights and ideas for what alternative hypotheses to explore.
Dissemination Phase
The final phase of data science is disseminating results, most commonly in the form of written reports such as internal memos, slideshow presentations, business/policy white papers, or academic research publications. The main challenge here is how to consolidate all of the various notes, freehand sketches, emails, scripts, and output data files created throughout an experiment to aid in writing.
Beyond presenting results in written form, some data scientists also want to distribute their software so that colleagues can reproduce their experiments or play with their prototype systems. For example, computer graphics and user interface researchers currently submit a video screencast demo of their prototype systems along with each paper submission, but it would be ideal if paper reviewers could actually execute their software to get a “feel” for the techniques being presented in each paper. In reality, it is difficult to distribute research code in a form that other people can easily execute on their own computers. Before colleagues can execute one’s code (even on the same operating system), they must first obtain, install, and configure compatible versions of the appropriate software and their myriad of dependent libraries, which is often a frustrating and error-prone process. If even one portion of one dependency cannot be fulfilled, then the original code will not be re-executable.
Similarly, it is even difficult to reproduce the results of one’s own experiments a few months or years in the future, since one’s own operating system and software inevitably get upgraded in some incompatible manner such that the original code no longer runs. For instance, academic researchers need to be able to reproduce their own results in the future after submitting a paper for review, since reviewers inevitably suggest revisions that require experiments to be re-run. As an extreme example, my former officemate Cristian Cadar used to archive his experiments by removing the hard drive from his computer after submitting an important paper to ensure that he can re-insert the hard drive months later and reproduce his original results.
Lastly, data scientists often collaborate with colleagues by sending them partial results to receive feedback and fresh ideas. A variety of logistical and communication challenges arise in collaborations centered on code and data sharing.
For further reading, check out my Ph.D. dissertation projects for examples of prototype tools that address the challenges mentioned in this post.





Join the Discussion (0)
Become a Member or Sign In to Post a Comment