The fruit fly’s neurological development is serving as a model for a networking algorithm that researchers say has the potential to obsolesce traditional methods for determining peer relationships in distributed networks. Without prior knowledge of how cells are connected, the fly’s developing nervous system allocates certain cells as leaders that provide direct connections with other nerve cells. That development process, the researchers say, is similar to conventional schemes used to manage distributed networks but is much simpler and more robust than anything humans have yet devised.
The lead scientist of the project to develop the new networking algorithm is Ziv Bar-Joseph, an associate professor of machine learning and computational biology at Carnegie Mellon University. Bar-Joseph says he became interested in biology-inspired computing while studying at Massachusetts Institute of Technology during the time of the initial sequencing of the human genome. “I was fascinated by the opportunities this information presented for improving human health and our understanding of the inner workings of cells in our body,” he says. “However, I also realized that the explosion of data coming out of biology would require the development of new computational methods to make sense of this data.”
More than a decade later, the need for sophisticated methods for dealing with increasingly large sets of biological data and even new types of information remains a salient characteristic of both computational biology and biology-inspired computing, with the former applying computational methods to biology and the latter drawing design inspiration from biological processes.
Following earlier work—in neural networking, for example—that focused on creating computing models developed from a high-level understanding of biological systems, researchers working today on biology-inspired computing projects study biological systems at a much lower level, deriving insight from the way molecular systems function. While this new generation of biology-inspired computing is relatively young, researchers in the field are developing computing approaches that already are having an impact on computer science.
But despite the promise of innovative, biological solutions to classic computing problems, the field of biology-inspired computing is filled with many challenges. One of the major issues facing the field is to develop new languages and methodologies so team members, having different backgrounds and expertise, will understand each other more effectively.
Biology-inspired computing is relatively young, but researchers in the field have developed computing approaches that already are having an impact on computer science.
Bar-Joseph says the issue of team communication is improving, especially with several new Ph.D. programs in computational biology enabling students to achieve fluency in both biology and computer science. However, despite the rise of dedicated graduate programs, computer scientists and biologists often take very different approaches to their work. “Finding a common problem that would be of interest to both sides is something that I view as one of the major challenges in this area,” says Bar-Joseph. “You want to work on cutting-edge computer science research while still advancing biology. It’s not trivial to find such problems and good collaborators to work with.”
A Fortunate Visit
The fruit fly research undertaken by Bar-Joseph and his colleagues—Noga Alon and Yehuda Afek of Tel Aviv University and Naama Barkai, Eran Hornstein, and Omer Barad of the Weizmann Institute of Science—emerged from what Bar-Joseph calls an “accidental event” during a visit to a colleague’s lab where a graduate student described research on the molecular mechanisms involved in cell fate determination. Bar-Joseph says the described research seemed to apply to a problem in networking, so the team set about designing the fruit fly project that eventually resulted in what the researchers say is a breakthrough algorithm for distributed networks.
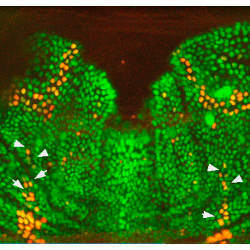
In networking, one method of creating a distributed framework is to allocate a set of leader nodes, sometimes called a maximal independent set (MIS), which can be used to communicate with the network’s other nodes. Every node in such a network is a leader or is connected to a leader, but the leaders are not interconnected. The researchers found that a similar arrangement occurs in the fruit fly, which uses tiny hairs for sensing. Each hair develops from a nerve cell called a sensory organ precursor (SOP), which connects to adjoining nerve cells but does not connect to other SOPs.
Computer scientists have developed several methods to determine how best to elect an MIS in distributed networks. One popular approach is to use a probabilistic method in which some nodes identify themselves as leaders on the basis of how many connections they have with other nodes. Nodes connected to these self-selected leaders take themselves out of the running in subsequent selection rounds. While this selection process is rapid, the network is inundated with messages because all nodes must continuously monitor how they are connected.
According to the researchers, the fruit fly’s cells have no information about how they are connected and no way to determine how many neighbors they have. As various cells self-select as SOPs, they send out chemical signals that inhibit neighboring cells from becoming SOPs. This process continues until cells are either SOPs or are neighbors of an SOP. The researchers say the probability that any cell will self-select as an SOP during the fly’s development increases not as a function of connections, as in the typical algorithm for distributed networks, but as a function of time.
The algorithm developed by Bar-Joseph and colleagues balances trying to reduce the number of potential collisions—neighboring nodes mistakenly electing themselves as leaders—against trying to quicken the selection of the MIS. To create this trade-off, the new algorithm relies on a time-based method that uses weighted randomization to decide whether a node will nominate itself as a leader node. Determining the probability of the randomization is tricky, the researchers say, because a high probability of self-selection means too many collisions and too many leader nodes. Conversely, a low probability means too few nodes would elect themselves.
Following the pattern of the fruit fly’s development, the algorithm varies that probability over time, starting low and increasing the likelihood that a node will self-select. “It turns out that if you have lots of neighbors, you would be selected early on, or connected to someone selected, whereas if you have few neighbors, it might take a bit longer,” says Bar-Joseph.
The researchers concede that, at this point, the new algorithm is slower than conventional methods for determining an MIS in a distributed network. But that speed shortfall, they say, is outweighed by efficiency, especially in the context of sensor networks where speed in establishing and maintaining an MIS is not the primary objective. The new algorithm is designed to operate efficiently with few assumptions about node location and with a minimal messaging overhead.
“In sensor networks, the nodes need to initially establish some backbone, which is what our algorithm does,” says Ziv Bar-Joseph. “For such sensors, energy efficiency is an important issue, which is why we believe our method should become the method of choice for these applications.”
Sensor networks, often used to monitor environmental conditions, typically consist of inexpensive devices that can be scattered across a large area. In these networks, there is no prior knowledge of where each sensor will be positioned or even whether all sensors will remain within range of all other sensors. “In sensor networks, the nodes need to initially establish some backbone, which is what our algorithm does,” says Bar-Joseph. “For such sensors, energy efficiency is an important issue, which is why we believe our method should become the method of choice for these applications.”
While the algorithm appears to be well suited to sensor networks, whether its creation will have implications for other areas of distributed computing remains to be seen. For his part, Bar-Joseph says he is hopeful that such biology-inspired ideas will help improve networking frameworks so they can operate more effectively in conditions that challenge today’s best fault-tolerant designs. “Biological systems are often distributed, both within and between cells,” he says. “As such, they face many of the same challenges faced by modern complex computer systems, so fault tolerance that is a major issue in large computer networks is an issue that we can study in these systems as well.”
Bar-Joseph and his colleagues are working on several new projects in this area, trying to determine how cellular failure can provide clues about which nodes in computer networks are best maintained with backups. He and his team are also working on trying to develop more effective methods for determining the appropriate connectivity of computer networks so they can withstand node failures and message corruption. Indeed, the field of biology-inspired computing appears to be full of possibilities for innovative projects that investigate real-life analogues to computing scenarios.
“As computer science matures, it will start to look outward much more than it does now and play a bigger role in several other areas of science and technology,” says Bar-Joseph. “We are still a rather young discipline, so most of the efforts are inward-looking, but this is slowly changing.”
Afek, Y., Alon, N., Barad, O., Hornstein, E., Barkai, N., and Bar-Joseph, Z.
A biological solution to a fundamental distributed computing problem, Science 331, 6014, Jan. 14, 2011.
Ater-Kranov, A., Bryant, R., Orr, G., Wallace, S., and Zhang, M.
Developing a community definition and teaching modules for computational thinking, Proceedings of the 2010 ACM Conference on Information Technology Education, Midland, MI, Oct. 79, 2010.
Gitter, A., Siegfried, Z., Klutstein, M., Fornes, O., Oliva, B., Simon, I., and Bar-Joseph, Z.
Backup in gene regulatory networks explains differences between binding and knockout results, Molecular Systems Biology, June 16, 2009.
Kephart, J. O.
Learning from nature, Science 331, 6018, Feb. 11, 2011.
Tero, A., Takagi, S., Saigusa, T., Ito, K., Bebber, D. P., Fricker, M. D., Yumiki, K., Kobayashi, R., and Nakagaki, T.
Rules for biologically inspired adaptive network design, Science 327, 5964, Jan. 22, 2010.





Join the Discussion (0)
Become a Member or Sign In to Post a Comment