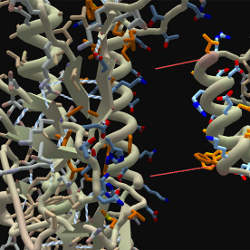
Scientists at the University of Washington had struggled for more than a decade to discover the structure of a protein that helps the human immunodeficiency virus multiply. Understanding its shape could aid them in developing drugs to attack the virus, but the scientists had been unable to decipher it. So they turned to colleagues who had developed Foldit, an online game that challenges players to rearrange proteins into their lowest-energy form, the form they would most likely take in nature.
Within three weeks the 57,000-plus players, most of whom had no training in molecular biology, had the answer, which was subsequently published in the journal Nature Structural and Molecular Biology. The University of Washington researchers say this was probably the first time a long-standing scientific problem had been solved using an online game.
Adrien Treuille, an assistant professor of computer science at Carnegie Mellon University, and one of the creators of Foldit, says the game is an example of a rather startling insight. “Large groups of non-experts could be better than computers at these really complex problems,” he says. “I think no one quite believed that humans would be so much better.”
There are many problems in science that computers, despite their capacity for tackling vast sets of data, cannot yet solve. But through the clever use of the Internet and social media, researchers are harnessing abilities that are innate to humans but beyond the grasp of machines—intuition, superior visual processing, and an understanding of the real world—to tackle some of these challenges. Ultimately, the process may help teach computers how to perform these tasks.
Computers can try to figure out the structures of proteins by rearranging the molecules in every conceivable combination, but such a computationally intensive task could take years. The University of Washington researchers tried to speed up the process using Rosetta@Home, a distributed program developed by University of Washington biochemistry professor David Baker to harness idle CPU time on personal computers. To encourage people to run the program, they created a graphical representation of the protein folding that acted as a screensaver. Unexpectedly, says Treuille, people started noticing they could guess whether the computer was getting closer to or farther from the answer by watching the graphics, and that inspired the idea for Foldit.
The game’s object is to fold a given protein into the most compact shape possible, which varies depending on how its constituent amino acids are arranged and on the attractive and repulsive forces between them. With a little practice, humans can use their visual intuition to move parts of the protein around on the screen into a shape that seems more suitable. Players’ scores are based on a statistical estimate of how likely it is that the shape is correct.
Treuille has since devised a new game, EteRNA, which reverses the process. In it, players are given shapes they are not allowed to change, and have to come up with a sequence of amino acids they think will best fit. A biochemistry lab at Stanford University then synthesizes the player-designed proteins. The biochemists can compare the structures that computer models predict with those the players create, and perhaps learn the reasons for the differences. Knowing the structure of a protein helps scientists understand its functions.
Since researchers launched Phylo in late 2010, more than 35,000 people have played the game, and they have improved approximately 70% of the alignments they have been given.
In some types of crowdsourcing, computers perform the preliminary work and then people are asked to refine it. For instance, Jerome Waldispuhl and Mathiue Blanchette, assistant professors of computer science at McGill University, have designed Phylo, a game that helps them align sequences of DNA to study genetic diseases. Geneticists can trace the evolution of genes by comparing them in closely related species and noting where mutations have changed one or more of the four nucleotides that make up DNA. Comparing three billion nucleotides in each of the 44 genomes of interest would require far too much computer time, so researchers use heuristics to speed up the computations, but in the process lose some accuracy.
Phylo shows humans some snippets of the computer’s solutions, with the nucleotides coded by color, and asks them to improve the alignments. The challenge is that the matches are imperfect, because of the mutations, and there can also be gaps in the sequence. Humans, Waldispuhl explains, have a much easier time than computers at identifying a match that is almost but not quite perfect. Since the McGill University researchers launched Phylo in late 2010, more than 35,000 people have played the game, and they have improved approximately 70% of the alignments they have been given.
See Something, Say Something
Like Phylo, many crowdsourcing projects rely on humans’ vastly superior image-processing capabilities. “A third of our brain is really wired to specifically deal with images,” says Luis von Ahn, associate professor of computer science at Carnegie Mellon. He developed the ESP Game, since acquired by Google, which shows a photo to two random people at the same time and asks them to type a word that describes what it depicts; the people win when they both choose the same word. As they play the game, they also create tags for the images. Scientists hope computers can apply machine learning techniques to the tags and learn to decipher the images themselves, but von Ahn says it could be decades before machines can match people. “Eventually we’re going to be able to get computers to be able to do everything humans can do,” he says. “It’s just going to take a while.”
In Duolingo, launched in late 2011, von Ahn relies on humans’ understanding of language to get crowds to do what computers do inadequately—translate text. Duolingo helps people learn a foreign language by asking them to translate sentences from that language to their own, starting with simple sentences and advancing to more complex ones as their skills increase. The computer provides one-to-one dictionary translations of individual words, but humans use their experience to see what makes sense in context. If one million people participated, von Ahn predicts they could translate every English-language page of Wikipedia into Spanish in just 80 hours.
Another skill humans bring to crowdsourcing tasks is their ability to notice unusual things and ask questions that were not part of the original mandate, says Chris Lintott, a researcher in the physics department at the University of Oxford. Lintott runs Zooniverse, a collection of citizen science projects, most of which ask people to identify objects in astronomical images, from moon craters to elliptical galaxies. In one project, a Dutch teacher using Galaxy Zoo to identify galaxies imaged by the Sloan Digital Sky Survey noticed a mysterious glow and wanted to know what it was. Astronomers followed up on her discovery, which is now believed to be the signature of radiation emitted by a quasar. Another Zooniverse project asks people to look through scanned pages of World War I-era Royal Navy logs and transcribe weather readings; climatologists hope to use the data to learn more about climate change. On their own, some users started keeping track of how many people were on a ship’s sick list each day, and peaks in the sick list numbers turned out to be a signature of the 1918 flu pandemic. “When people find strange things, they can act as an advocate for them,” Lintott says. If computers cannot even identify the sought-after information, they are hardly going to notice such anomalies, let alone lobby for an answer.
Payoffs For Players
Designing crowdsourcing projects that work well takes effort, says von Ahn, who tested prototypes of Duolingo on Amazon Mechanical Turk, a service that pays people small amounts of money for performing tasks online. Feedback from those tests helped von Ahn make the software both easier to use and more intuitive. He hopes Duolingo will entice users to provide free translation services by offering them value in return, in the form of language lessons. Paying for translation, even at Mechanical Turk’s low rates, would be prohibitively expensive.
Lintott agrees the projects must provide some type of payoff for users. There is, of course, the satisfaction of contributing to science. And for Zooniverse users, there is aesthetic pleasure in the fact that many galaxy images are strikingly beautiful. For Phylo and Foldit users, there is the appeal of competition; the score in Foldit is based on a real measure of how well the protein is folded, but the score is multiplied by a large number to make it like players’ scores in other online games.
If one million people participated in Duolingo, Luis von Ahn predicts they could translate every English-language page of Wikipedia into Spanish in just 80 hours.
The University of Washington’s Center for Game Science, where Foldit was developed, is exploring the best ways to use what center director Zoran Popović calls “this symbiotic architecture between humans and computers.” He hopes to use it to harness human creativity by, for instance, developing a crowdsourcing project that will perform the efficient design of computer chips. That is not as unlikely as it sounds, Popović says. The non-experts using Foldit have already produced protein-folding algorithms that are nearly identical to algorithms that experts had developed but had not yet published.
The challenge, Popović says, is to figure out how to take advantage of this symbiosis. “It’s just a very different architecture to compute on, but it’s highly more productive.”
Further Reading
Cooper, S., et al.
The challenge of designing scientific discovery games, 5th International Conference on the Foundations of Digital Games, Monterey, CA, June 1921, 2010.
Khatib, F., et al.
Algorithm discovery by protein folding game players, Proceedings of the National Academies of Science 108, 47, November 22, 2011.
Nielsen, M.
Reinventing Discovery: The New Era of Networked Science, Princeton University Press, Princeton, NJ, 2011.
Smith, A. M., et al.
Galaxy Zoo supernovae, Monthly Notices of the Royal Astronomical Society 412, 2, April 2011.
Von Ahn, L. and Dabbish, L.
Designing games with a purpose, Communications of the ACM 51, 8, August 2008.



Join the Discussion (0)
Become a Member or Sign In to Post a Comment